- This event has passed.
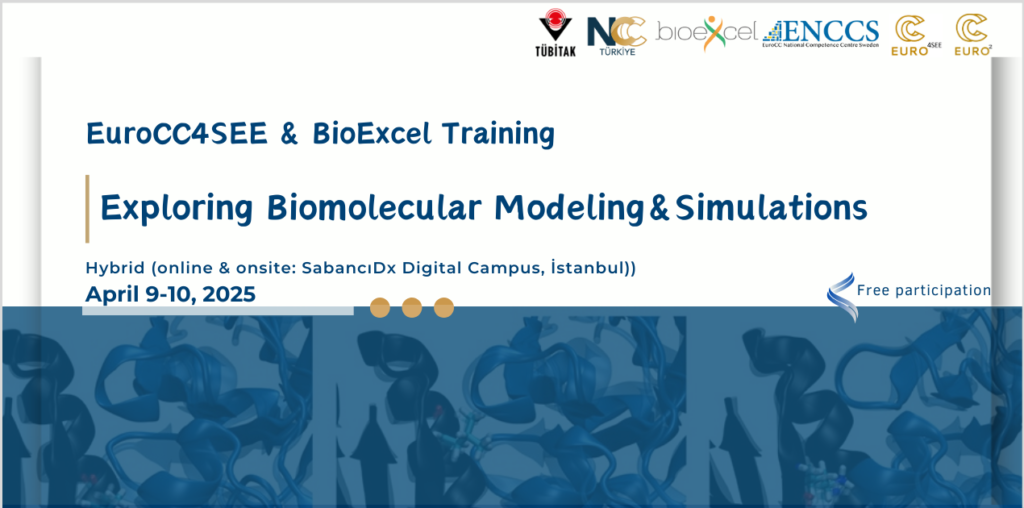
Exploring Biomolecular Modelling and Simulations
Apr 9 • 09:00 – Apr 10 • 17:00 Europe/Istanbul
Registration Deadline: 24 March 2025
Format: Hybrid (on-site & online)
Prerequisites:
-Experience in a Linux environment, jupyter notebook, Python, Conda, and BioBB library.
-Familiarity with High-Performance Computing (HPC) systems is required for participation in the hands-on sessions.
-The training is primarily designed for participants from academia (with at least a graduate degree), industry, or public administration who are using or planning to use biomolecular modeling and simulation in their research.
For the event programme see the timetable section.
The training consists of four sessions:
1-) Integrative modeling of biomolecular complexes with HADDOCK
Tools, libraries, and applications used:
HADDOCK3: https://github.com/haddocking/haddock3
HADDOCK web server: https://wenmr.science.uu.nl
HADDOCK tutorials: https://bonvinlab.org/education
Learning outcomes:
Getting insights into the field of integrative modeling and more specifically into HADDOCK.
Gaining hands-on experience in the modeling of biomolecular complexes with HADDOCK
Instructor: Alexandre M.J.J Bonvin
2-) Molecular Dynamics Simulations – GROMACS
Tools, libraries, and applications used:
GROMACS: https://www.gromacs.org/
VMD: https://www.ks.uiuc.edu/Development/Download/download.cgi?PackageName=VMD
Tutorial: http://www.mdtutorials.com/gmx/index.html, https://tutorials.gromacs.org/
Learning outcomes:
Understanding the scope of MD simulations and the key considerations before setting up a simulation.
Hands-on experience on MD simulation using GROMACS.
Instructor: Nandan Haloi, PhD
3-) Interoperable and Reproducible Biomolecular Simulation Workflows using BioExcel Building Blocks (BioBB)
Tools, libraries, and applications used: Python, Conda, and BioBB library.
Information about the library, installation protocols, and dependencies can be found here: https://mmb.irbbarcelona.org/biobb/;
All the tutorials are available here: https://mmb.irbbarcelona.org/biobb/workflows
Learning outcomes: Learn how to build FAIR (Findable, Accessible, Interoperable, and Reusable/Reproducible) biomolecular simulation workflows using the BioBB library.
Instructor: Adam Hospital
4) Fundamentals and Practical Use Cases of Free Energy Calculations with PMX
Tools, libraries, and applications used: Conda and GROMACS.
The installation protocol and dependencies are documented here (http://pmx.mpibpc.mpg.de/Tutorial2024/Coimbra_2024/ligand_tutorial.html).
All the tutorials are available here: http://pmx.mpibpc.mpg.de/tutorial.html
Learning outcomes:
(i) the basics of free energy calculations
(ii) their use in predicting protein-ligand binding free energies with PMX and GROMACS, with direct relevance to drug design.
Instructor: Sudarshan Behera
Contact:ncc@ulakbim.gov.tr
Notes:
Participants will use their own laptop or computer for the hands-on sessions.
Before the training, participants will receive instructions on how to install the software and configure the environment.
Hybrid format
The training consists of lectures and hands-on sessions. All registered participants will be able to follow the lectures; however, the hands-on sessions will have a limited capacity of 100 people (40 face-to-face particpants and 60 online participants). Based on the registration form, participants will be selected by considering their experience, area of research, and qualifications. Participants should meet the qualification requirements of the hands-on sessions. The acceptance notification will be sent at least 10 days before the event. Selected participants (for the hands-on sessions) will receive a temporary user account for the hands-on training.
Please note that the allocated compute time should only be used for running the hands-on examples and that only limited technical support will be provided to online participants!
A certificate of attendance will be given only to participants who will follow a minimum of 70% of the whole duration of the course and will be signed in with full name.
Regulations
Due to EuroCC2 regulations, we CAN NOT ACCEPT generic or private email addresses. Please use your official university or company email address for registration.
This training is for users who live and work in the European Union or a country associated with Horizon 2020. You can read more about the countries associated with Horizon2020 HERE.